A divalent interaction between HPS1 and HPS4
Hermansky-Pudlak syndrome (HPS) is a group of rare autosomal recessive disorders characterized by oculocutaneous albinism, bleeding tendency, and sporadic pulmonary fibrosis, granulomatous colitis, or infections. Nine genes responsible for HPS have been identified in humans. HPS-1 is the most severe subtype with a prevalence of approximately 1/1800 in northwestern Puerto Rico due to a founder mutation in the HPS1 gene. Mutations in HPS genes affect the biogenesis of lysosome-related organelles such as melanosomes in melanocytes and platelet-dense granules.
Two of these genes (HPS1 and HPS4) encode the proteins HPS1 and HPS4, which assemble to form a complex called Biogenesis of Lysosome-related Organelle Complex 3 (BLOC-3). We report the identification of interacting regions in HPS1 and HPS4 required for the formation of this complex. Two regions of HPS1, spanning amino acids 1-249 and 506-700, are required to bind HPS4.
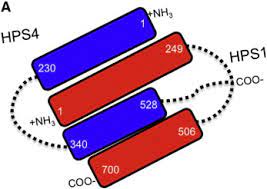
The middle part of HPS1 (residues 250–505) is not required for this interaction. Other interaction studies have shown that the N-termini of HPS1 and HPS4 interact with each other and that a discrete region of HPS4 (residues 340 to 528) interacts with the N- and C-termini of the HPS1 protein .
Several missense mutations found in HPS-1 patients did not affect the interaction with HPS4, but some mutations involving regions interacting with HPS4 caused HPS1 instability. These observations expand our understanding of BLOC-3 assembly and represent an important first step in identifying the domains responsible for the biogenesis of lysosome-like organelles.
An HPS1 isoform encoded by a transcript lacking exon 9 interacts with HPS4 The central region of the HPS4 protein interacts in a dual manner, either with the N- or C-terminal end of HPS1 The interaction regions of HPS1 and HPS4 exist in soluble and membrane-associated form Missense mutations in interacting regions of HPS1 alter its stability The membrane association of HPS1 and HPS4 depends on specific regions of these proteins
