A novel immune biomarker IFI27
Host response biomarkers can accurately distinguish between influenza and bacterial infection. However, the published biomarkers require the measurement of many genes, which makes their implementation in clinical practice difficult. This study aims to identify a single gene biomarker with high diagnostic accuracy equivalent to multigene biomarkers.
In this study, we combined integrated genomic analysis of 1071 individuals with in vitro experiments using well-established infection models.
We identified a single gene biomarker, IFI27, which had a high prediction accuracy (91%) equivalent to that obtained by multigene biomarkers. In vitro studies showed that IFI27 was upregulated by TLR7 in plasmacytoid dendritic cells, antigen-presenting cells that responded to influenza virus rather than bacteria.
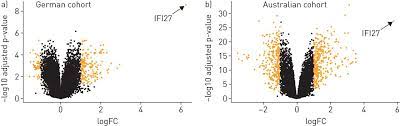
In vivo studies confirmed that IFI27 was expressed in influenza patients but not in bacterial infections, as demonstrated in multiple patient cohorts (n=521). In a large prospective study (n=439) of patients with undifferentiated respiratory disease (etiologies included viral, bacterial, and non-infectious conditions), IFI27 showed a diagnostic accuracy (AUC) of 88% and a specificity of 90 % to distinguish between influenza and bacterial infections.
IFI27 represents a significant advance in overcoming a translational barrier in the application of genomic testing in the clinical setting; its implementation can improve the diagnosis and management of respiratory infections.
Acute respiratory tract infections are the most common causes of morbidity and mortality from infectious diseases worldwide (WHO - 2014). Viruses and bacteria are the main causes of respiratory infections. Among viruses, the influenza virus causes the highest morbidity and mortality rates. Clearly differentiating between influenza virus and bacteria is the first key step in the management of respiratory infections.
since the initial treatment (antiviral versus antibiotic) is mainly guided by the etiology. However, it is often difficult to differentiate between influenza and bacterial infections on clinical grounds because the two infections share similar clinical features (eg, cough, dyspnoea, and fever).
Screening for pathogens (eg virus detection by PCR) is an important step towards clarifying the diagnosis. Although important, virus detection alone is insufficient to guide patient management. Like most respiratory viruses, the influenza virus produces a broad clinical spectrum. In some patients, the virus acts as a bystander and its presence is unrelated to the disease presented.
In others, the virus is directly responsible for the patient's symptoms. Clinical decision-making requires determining whether the virus is a bystander or the cause of the presenting symptoms, as antiviral therapy (eg, oseltamivir) should only be given to people in whom symptoms are severe and directly caused by the virus [2–4]. Currently, there is no laboratory test to distinguish between detection of a virus and an "active infection".
